Shon Kurian George
Spatial Transcriptomics Pipeline Development
Image-based cell profiling (cypro : R package)
Edinburgh, United Kingdom & Kerala, India · Reachable on Email/LinkedIn · Open to Collaboration
About
Bioinformatician with an MSc in Bioinformatics from the University of Edinburgh shipping reproducible analyses and tools with an interest in providing interactive, easy-to-use applications. Skilled in R, Python, Bash Scripting and building bioinformatic pipelines.
Skills
- R / Shiny, Python, bash scripting
- Spatial transcriptomics (SPATA2, Seurat)
- Git/GitHub, Docker, AWS EC2, Nextflow, MySQL
- Excel, Vim, Ollama, Open WebUI
Work Experience
cypro R Package
R Package Developer · Nov 2024 – Present
Developing an R package to streamline image-based cell profiling workflows by integrating data from diverse platforms including CellProfiler and CellTracker. Enhanced package usability by implementing intuitive Shiny-based interfaces that allow detailed specification down to individual well-level configurations, significantly simplifying user interaction.
Refined S4 class objects within cypro to ensure accurate data integration from imaging platforms, enabling comprehensive cell movement analyses. Incorporated proactive user warnings and notifications within the Shiny application, minimising user errors, safeguarding data integrity, and enhancing the overall user experience.
View GitHub repo
10x Visium Spatial Transcriptomics Pipeline to Analyse miRNA Expression Patterns
MSc Bioinformatics Dissertation · Mar 2024 - Sep 2024
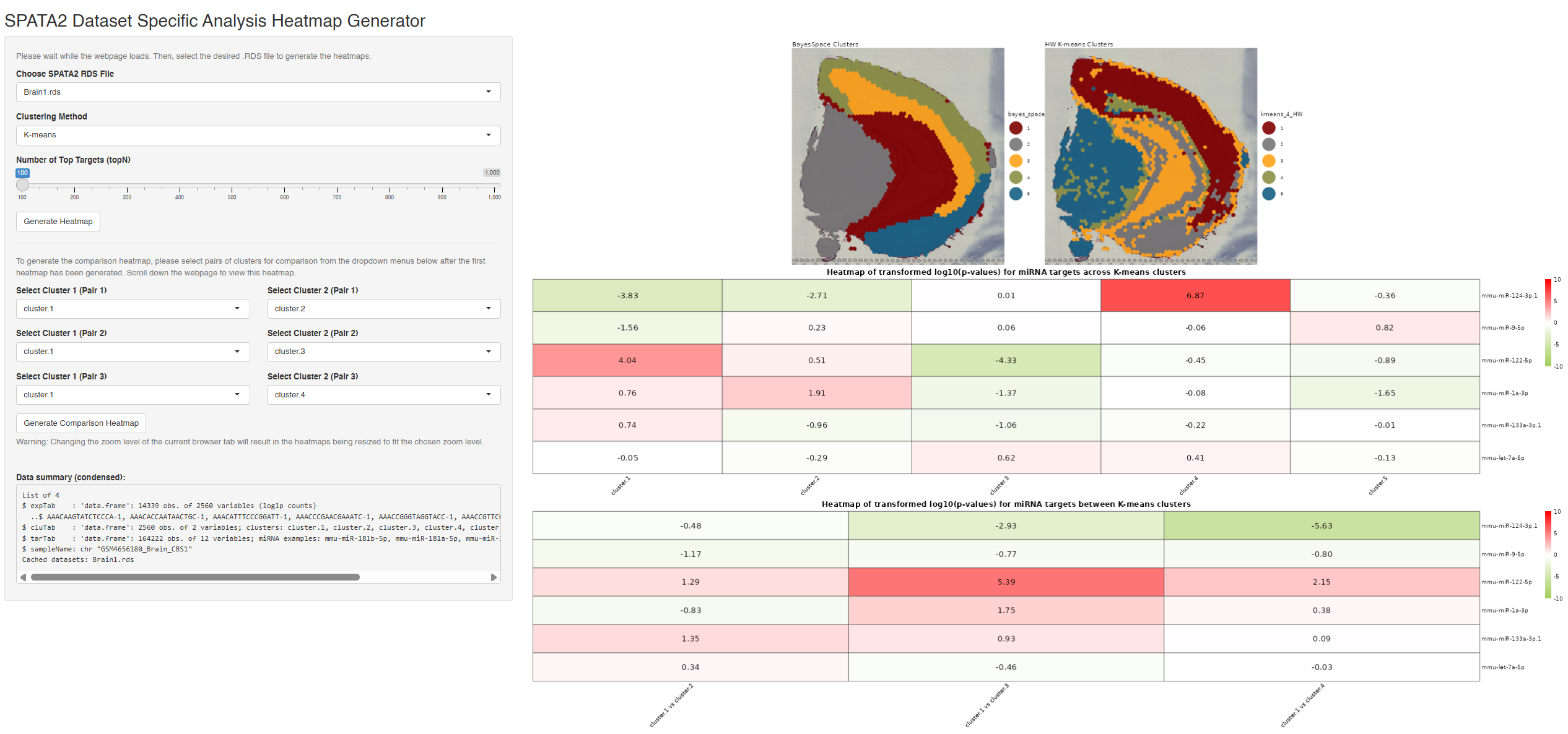
Developed an end-to-end pipeline to investigate whether miRNAs act as guardians of gene expression at cell-type boundaries. Utilised SPATA2 and Seurat frameworks to process datasets into analysis-ready form, then performed BayesSpace and Hartigan-Wong K-means clustering to segregate cell types.
Conducted TargetScan miRNA target analysis across 8 datasets (brain, heart, liver), detecting tissue-specific miR-124 (brain) and miR-1 (heart) with high confidence. Identified neighbouring clusters showing high and low target expression for both miRNAs. Built an accompanying Shiny app to interactively view the analysis results (SPATA2 Shiny App).
View GitHub repo
Interactive Applications (Hosted on AWS EC2)
👀 New Interactive Application 🍳
🚧 In Development
Stay tuned for updates!
Projects
Detailed Collaborative Machine Learning Project on Modifiable Risk Factors Linked to Dementia
Built a polynomial regression model using the SHARE (Survey of Health, Ageing and Retirement in Europe) dataset to predict cognitive scores and identify modifiable risk factors accounting for 40% of dementia cases worldwide. Cross-referenced the model findings with the Lancet Commission Population Attributable Factor (PAF) framework to analyse 7 key interventions, revealing mental health, physical activity, social engagement, and education as critical lifestyle interventions for reducing dementia risk.
Read the report
Analysis and Critique of Automeris io moth de novo Genome Assembly and Annotation by Skojec et al. and assembly using wtdbg2.
Executed a critical evaluation of the genome assembly workflow performed by Skojec et al. and compared assembly quality between Hifiasm (N50: 15.78 Mb) and wtdbg2 (N50: 1.1 Mb). Demonstrated the superior performance of Hifiasm with 98.4% completeness and a 490 Mb assembly across only 600 contigs (vs 3,362).
Read the analysis